In this blog post, I will introduce two configuration-driven Azure Data Factory pipeline patterns I have used in my previous projects, including the Source-Sink pattern and the Key-Value pattern.
The Source-Sink pattern is primarily used for parameterising and configuring the data movement activities, with the source location and sink location of the data movement configured in a database table. In Azure Data Factory, we can use Copy activity to copy an data entity (database table or text-based file) from a location to another, and we need to create an ADF dataset for the source and an ADF dataset for the sink. It can easily turn into a nightmare when we have a large number of data entities to copy and we have to create a source dataset and a sink dataset for each data entity. With the Source-Sink pattern, we only need to create a generic ADF dataset for each data source type, and dynamically refer to the data entity for copy with the source and sink settings configured in a separate configuration database.
The Key-Value pattern is used for configuring the settings of activities in Azure Data Factory, such as the notebook file path of a Databricks Notebook activity or the url of a logic app Https event. Instead of hard-coding the settings in ADF pipeline itself, the Key-Value pattern allows to configure the activity settings in a database table and to load the settings and dynamically apply them to the ADF activities at runtime.
In the rest of this blog post, I will introduce the implementation of those two patterns in Azure Data Factory.
Source-Sink Pattern
To implement the Source-Sink pattern in Azure Data Factory, the following steps need to be followed:
- Step 1 – create data movement source-sink configuration table
- Step 2 – create generic source and sink datasets
- Step 3 – use Lookup activity to fetch the source-sink settings of the data entities for copy from the configuration table
- Step 4 – use ForEach activity to loop through the data entities for copy
- Step 5 – add a generic Copy activity inner the ForEach activity
Step 1 – create data movement source-sink configuration table
Create a configuration database (an Azure SQL database will be a good option), and create a table with columns to define data movement category, location for source and location for sink.

Along with the configuration table, we need to create a stored procedure for the Lookup ADF activity to retrieve the source-sink settings of the data entities for specified copy category.

Step 2 – create generic source and sink datasets
Create generic source and sink datasets. To make the datasets generic, we need to create a number of dataset parameters to define the location of the table or file referred by the dataset. Those parameters will take the values fetched from the configuration database table at runtime.

On the Connection tab of the generic dataset, we need to specify the table name or file container/path/name using the dataset parameters we just defined.

Step 3 – use Lookup activity to fetch the source-sink settings
Add a Lookup activity and set to use the stored procedure we created earlier for retrieving the source-sink settings of all the data entities for copy in a specified category.

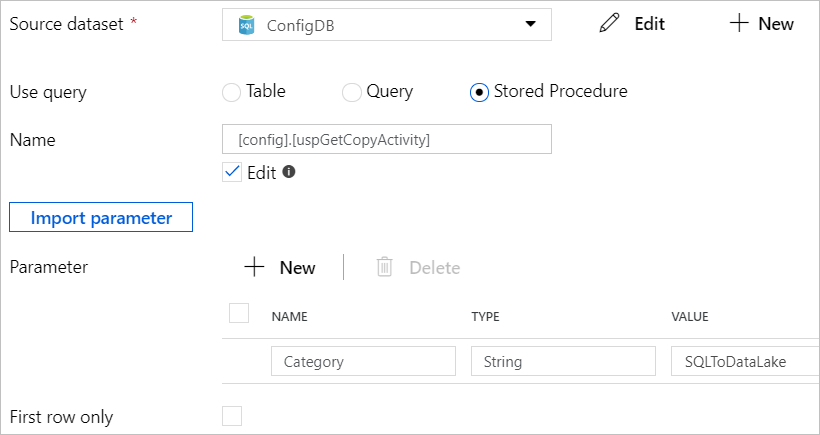
The output of the Lookup activity will contains source-sink settings of all the data entities in the specified category defined in the configuration database table.

Step 4 – use ForEach activity to loop through the data entities for copy
Add and chain a ForEach activity after the Lookup activity, and set the Items property of the ForEach activity as the output of the Lookup activity, @activity(‘Lookup Configuration’).output.value, so that the ForEach activity will loop through the source-sink settings of all the data entities fetched from the configuration database table.

Step 5 – add a generic Copy activity inner the ForEach activity
Inner the ForEach activity, add the Copy activity with the Source and Sink pointing to the generic source dataset and sink dataset we created earlier and pass the corresponding source-sink settings of current data entity item, such as @item().Sink_Item for the file name of the sink.

At this point, the source-sink pattern is all set. When the pipeline runs, a copy channel is created and executed for each data entity defined in the configuration database table.

Key-Value Pattern
To implement the Key Value pattern in Azure Data Factory, the following steps need to be followed:
- Step 1 – create activity settings key-value configuration table
- Step 2 – create stored procedure to transform the key-value table
- Step 3 – use Lookup activity to retrieve activity settings from configuration table
- Step 4 – apply settings on ADF activities
Step 1 – create activity settings key-value configuration table
Create a database table with columns defining pipeline name, activity type, activity name, activity setting key, activity setting value.

Step 2 – create stored procedure to transform the key-value table
The ADF Lookup activity is not working in the key-value type dataset. Therefore, we need to convert the key-value table into a single data row with column header as the activity setting key and the column value as the activity setting value.

We can use T-SQL PIVOT operator to convert the key-value table into the single row table. However, as the number of records in the key-value table is not fixed, we cannot specify the keys for pivot. Instead, we need to fetch the keys from the key-value table first and author the PIVOT operation in dynamical sql. The snapshot below shows the stored procedure I have created for convert the key-value table.

Step 3 – use Lookup activity to retrieve activity settings from configuration table
In the Azure Data Factory pipelines you are creating, at the start of the activities chain, Add a Lookup activity and use the stored procedure created earlier to retrieve the key-value table and convert it into the single row format to be readable for the Lookup activity. On the Lookup activity, tick the ‘First row only’ on as the result of the Lookup activity will be the single row dataset.
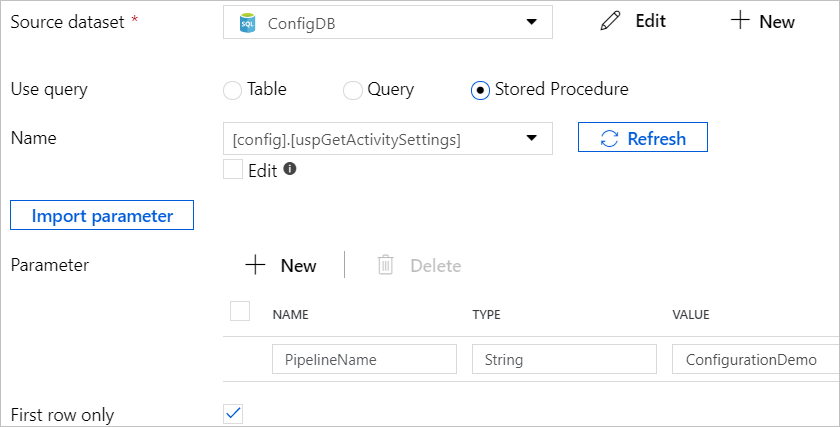
The snapshot below shows what the output of the Lookup activity looks like.

Step 4 – apply settings on ADF activities
From the output of the Lookup activity, the other ADF activities chained after can access the value of an activity setting as @activity(‘{lookup activity name}’).output.firstRow.{column name for the key}.


































You must be logged in to post a comment.